EBS-BA 25.09: Breeding Analytics Known Limitations
9 October, 2025
⚠️Breeding Analytics Known Limitations
These are known tool limitations. Workarounds are available, and fixes are scheduled for upcoming releases to improve reliability and enhance the user experience.
ARM (Single-Occurrence)
- Slow loading (~1 min) when selecting occurrences; performance depends on data size and network.
- Some jobs remain stuck in “Processing.”
Workaround: Run not more than 20 traits or jobs per analysis request. Fix in next sprint.
ARM (MOA)
- “Void” jobs can still be included in MOA results.
- MOA error may appear if weight values are blank or missing.
Molecular Data Analysis(MDA)
- Datasets with approximately 500 samples by 300 markers may load slowly or fail to start due to no background processing.
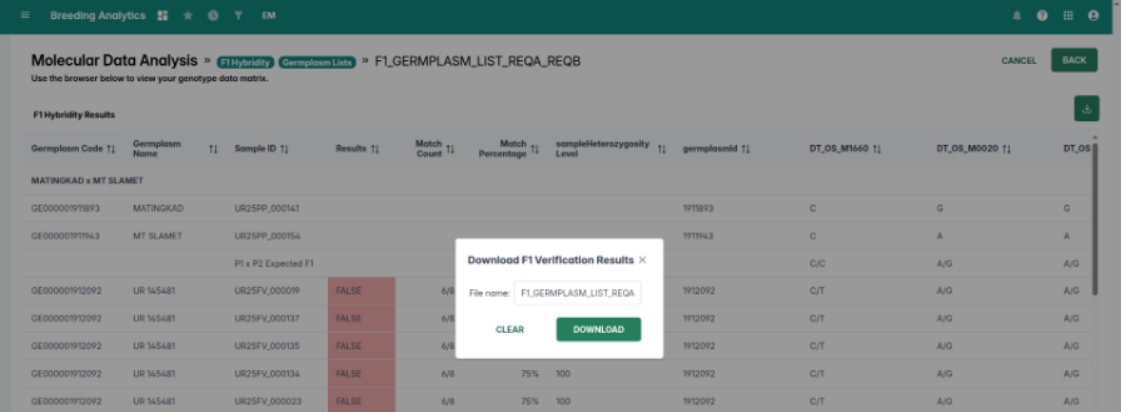
Planned: Background job handling and progress updates coming this year. - F1 Verification in MDA may show a 424 Failed Dependency error due to authorization key issues.
SDM-API
- Missing “entry type” or “entry role” fields are not yet validated during upload.
Fix: Field checks and defaults scheduled for next sprint.




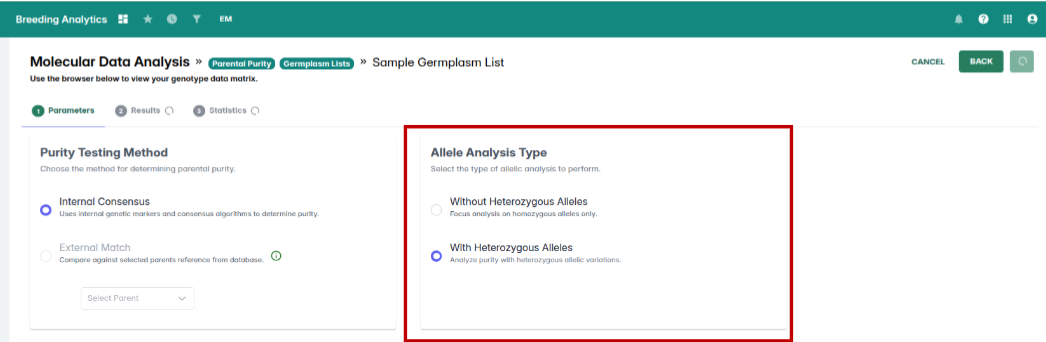
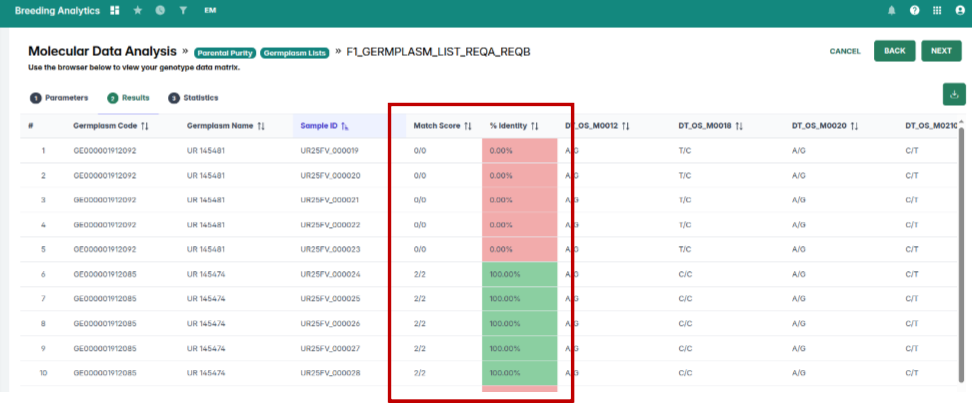
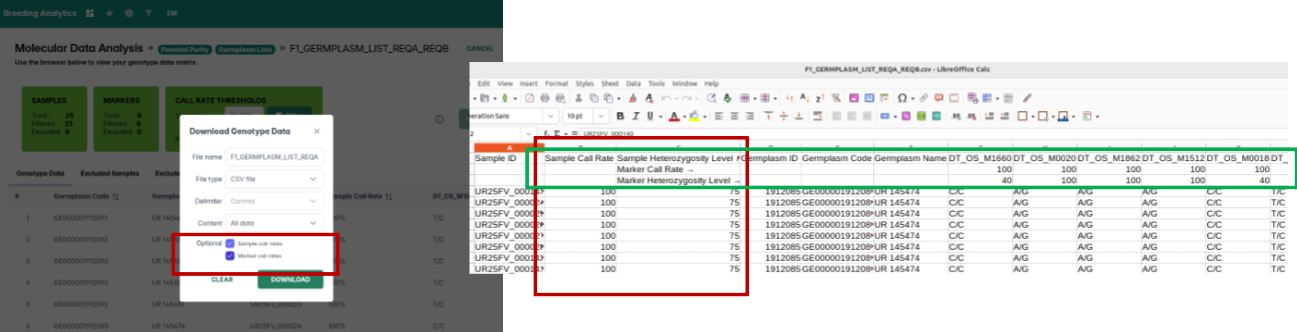
 Inconsistencies between the View QC/QA Data and Parental Purity tools have been resolved. Parental purity results now display correctly based on the germplasm list you select, ensuring aligned and dependable outputs.
Inconsistencies between the View QC/QA Data and Parental Purity tools have been resolved. Parental purity results now display correctly based on the germplasm list you select, ensuring aligned and dependable outputs.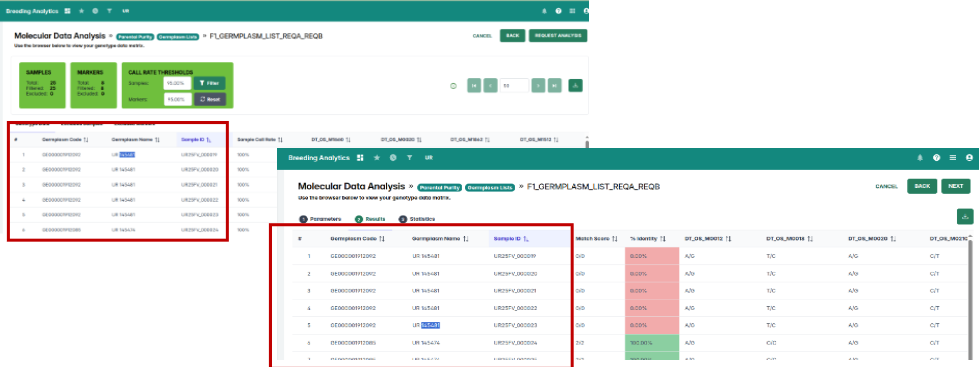

 Core Breeding Known Limitations
Core Breeding Known Limitations